microRNAs – function & biogenesis
What Are microRNAs?
MicroRNAs (miRNAs) are a class of small, non-coding RNA molecules typically 18–24 nucleotides in length. They play a crucial role in the post-transcriptional regulation of gene expression by base-pairing with complementary sequences, primarily within the 3′ untranslated regions (3′UTRs) of target messenger RNAs (mRNAs). This interaction leads to gene silencing, which can occur through mRNA cleavage, destabilization via deadenylation (shortening of the poly(A) tail), or translational repression.
Discovery and Conservation
miRNAs were first discovered in the nematode Caenorhabditis elegans in the early 1990s, and their significance in gene regulation has since been recognized across a wide range of organisms. To date, more than 2,500 mature miRNAs have been annotated in the human genome, many of which are highly conserved across animal species, highlighting their evolutionary and functional importance.
Nuclear Processing: From Pri-miRNA to Pre-miRNA
The biogenesis of miRNAs begins in the nucleus, where miRNA genes are transcribed by RNA polymerase II into long primary transcripts known as primary miRNAs (pri-miRNAs). These transcripts typically contain one or more stem-loop structures and are capped and polyadenylated like protein-coding mRNAs. The microprocessor complex, composed of the RNase III enzyme Drosha and its cofactor DGCR8 (DiGeorge syndrome critical region 8), recognizes and cleaves the pri-miRNA to produce a ~70-nucleotide precursor miRNA (pre-miRNA) with a characteristic hairpin structure.
Cytoplasmic Processing and RISC Loading
The pre-miRNA is then transported from the nucleus to the cytoplasm by the export receptor Exportin-5 in a Ran-GTP-dependent manner. In the cytoplasm, the RNase III enzyme Dicer further processes the pre-miRNA into a ~22-nucleotide miRNA duplex. One strand of this duplex, referred to as the guide strand, is loaded into the Argonaute-containing RNA-induced silencing complex (RISC), while the other strand (the passenger strand) is typically degraded.
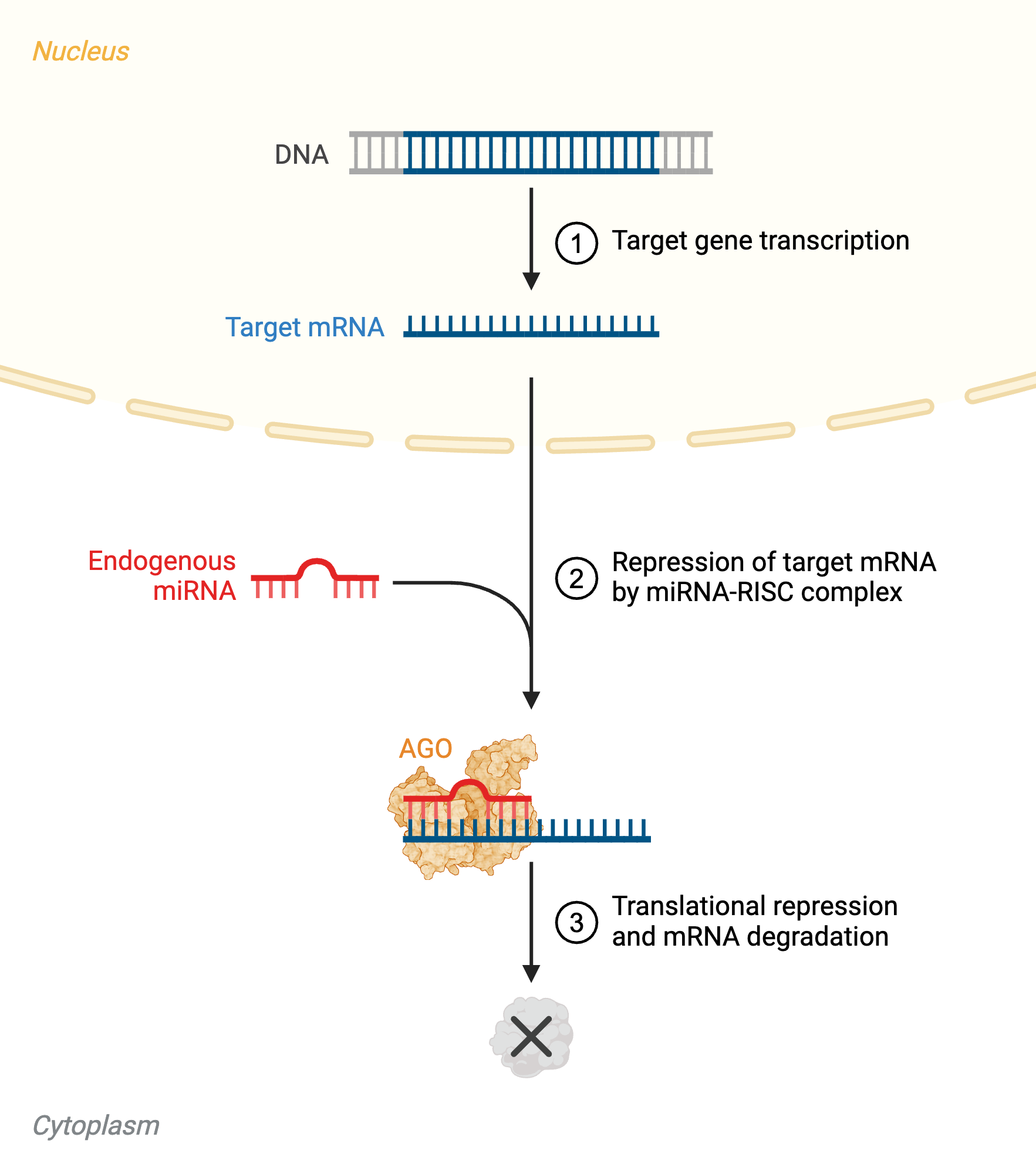
Target Recognition and Gene Silencing
The mature, single-stranded miRNA within RISC guides the complex to target mRNAs through partial base-pairing, primarily involving the so-called “seed region”—nucleotides 2–8 from the 5′ end of the miRNA—which plays a central role in target recognition. Perfect complementarity between the miRNA and its target is not required, allowing a single miRNA to regulate dozens to hundreds of different mRNA transcripts. This broad targeting capacity makes miRNAs key regulators of diverse cellular processes, including development, differentiation, apoptosis, and metabolism.
Recognition by the Nobel Prize
The importance of small RNAs in post-transcriptional gene regulation and their relevance to human health and disease has been recognized by two Nobel Prizes in Physiology or Medicine. In 2006, the award honored the discovery of RNA interference (RNAi), a gene-silencing mechanism involving small RNAs such as miRNAs and siRNAs. More recently, in 2024, the Nobel Prize recognized breakthroughs in understanding how cellular RNA regulation—including that mediated by small RNAs—contributes to health and disease, highlighting the growing clinical significance of RNA-based mechanisms.
microRNAs as Biomarkers
Beyond their role in gene regulation, microRNAs have emerged as promising biomarkers for a wide range of diseases. Their remarkable stability in body fluids such as blood, cerebrospinal fluid, and urine—combined with their tissue-specific expression patterns—makes them well-suited for non-invasive diagnostic and prognostic applications. Altered levels of circulating miRNAs have been associated with various pathological conditions, including cancer, cardiovascular disease, liver injury, and neurological disorders. Because miRNAs can reflect early molecular changes in disease processes, they hold particular value for early detection, monitoring of disease progression, and assessment of therapeutic response. Ongoing research and technological advances continue to support the development of miRNA-based assays for clinical use, highlighting their potential to complement or even surpass traditional biomarkers in sensitivity and specificity.
Read more: microRNAs & biomarkers
BioRender (2024). Mechanism of Action of miRNAs in mRNA Repression.
https://app.biorender.com/biorender-templates/figures/all/t-674dc0b4c5faa56c73079877-mechanism-of-action-of-mirnas-in-mrna-repression
You are currently viewing a placeholder content from YouTube. To access the actual content, click the button below. Please note that doing so will share data with third-party providers.
More Information






