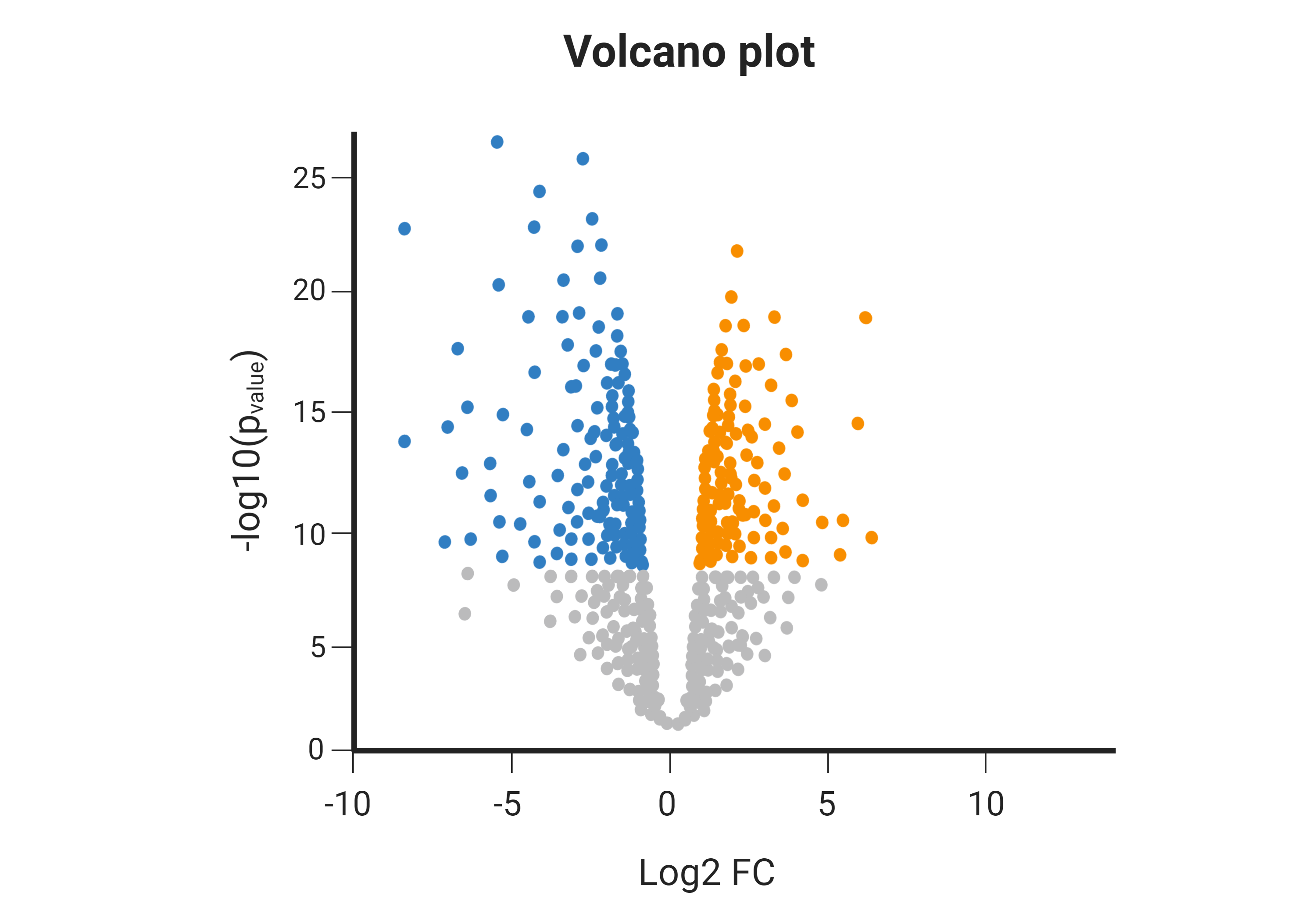
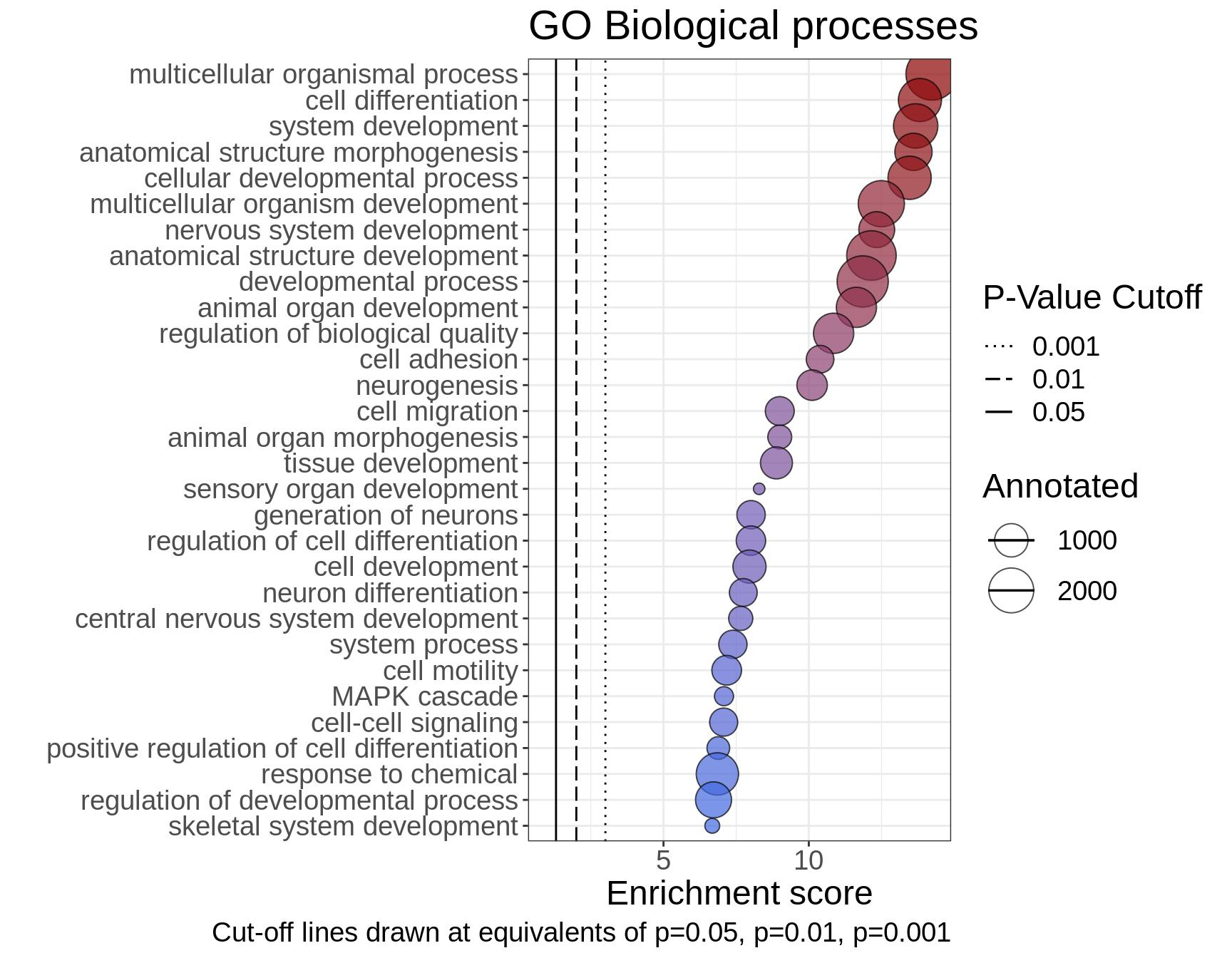
RNA sequencing (RNA-seq) services
We offer RNA sequencing services for
- messenger RNA (mRNA)
- long non-coding RNA (lncRNA), and
- circular RNA
analysis in any biological sample including cells, tissues, biofluids and extracellular vesicles (EVs).
We understand that RNA-sequencing data analysis can be challenging. Therefore, we perform full bioinformatic analysis with publication-ready reports to complement all our wet-lab services and consult our customers before and after each project.
Our 3’end RNAseq and whole transcriptome sequencing service is the preferred choice for a number of applications, such as:
- RNA biomarker discovery in cells, tissues, and biofluids
- Study of disease mode-of-actions
- Study of drug mode-of-action
Our RNA-seq services go hand-in-hand with our
- RT-qPCR services to further verify and validate NGS findings,
- small RNA-seq services to profile microRNA and mRNA expression to perform multiomic analysis
- Extracellular vesicle (EV) isolation and characterization services.
3’end and whole transcriptome sequencing (WTS) to target
- mRNA,
- long non-coding RNA,
- and circular RNAs
Compatible with cells, tissues, EVs/exosomes, and biofliuds.
benefits
What we offer: our service includes total RNA extraction, RNA quality control, library preparation, sequencing, quality control, and full statistical data analysis. For some applications (e.g. circular RNAs) we pretreat RNA to maximize yield and data quality.
Flexibility: The tasks that are taken over by TAmiRNA can be adjusted to customer requirements. We can conduct the entire RNA sequencing analysis or support customers with individual tasks such as library preparation, Illumina sequencing, or bioinformatic data analysis.
Best-in-class data analysis: we provide interactive data analysis reports that are ready to use for internal documentation, publications, and presentations. We have developed pipelines for mRNA, long non-coding RNA and circular RNA analysis.
Experience: we have experience in analyzing RNA sequencing data from thousands of samples including cells, tissue, nematodes, flys, whole-blood, cell-free biofluids, and extracellular vesicles/exosomes. We have worked with multiple species, including humans, mice, rats, horses, cows, pigs, and many others. Contact us for a free consultation.
service requirements
Using our optimized protocols we can deal with very low RNA inputs (<500 pg) obtained from cells or tissues.
We like to be challenged. In order to inquire about the possibility of analyzing biofluids, extracellular vesicles, or other challenging samples please get in touch with us.
service description
RNA sequencing includes several steps, which can all be performed at TAmiRNA:
- Total RNA extraction: we have protocols for cells, tissues, biofluids, and EVs. Leftover total RNA can be returned to customers for follow-up experiments.
- Total RNA quality control: concentration and integrity using highly sensitive and specific fluorescence-based methods
- RNA sequencing library preparation: we prepare high-quality 3′ mRNA-Seq libraries, which are ideal for gene expression analysis. Just one fragment per transcript is produced, which means that no length normalization is required. This allows more accurate determination of gene expression values and renders QuantSeq the best and most cost-effective alternative to microarrays and conventional RNA-Seq in gene expression studies.
- NGS analysis: we can access a wide-range of Illumina instruments (HiSeq, NextSeq, NovaSeq), which allows us to pick the most efficient solution for each project.
- Data analysis: we provide everything from raw data to fully analyzed data reports depending on customer needs.
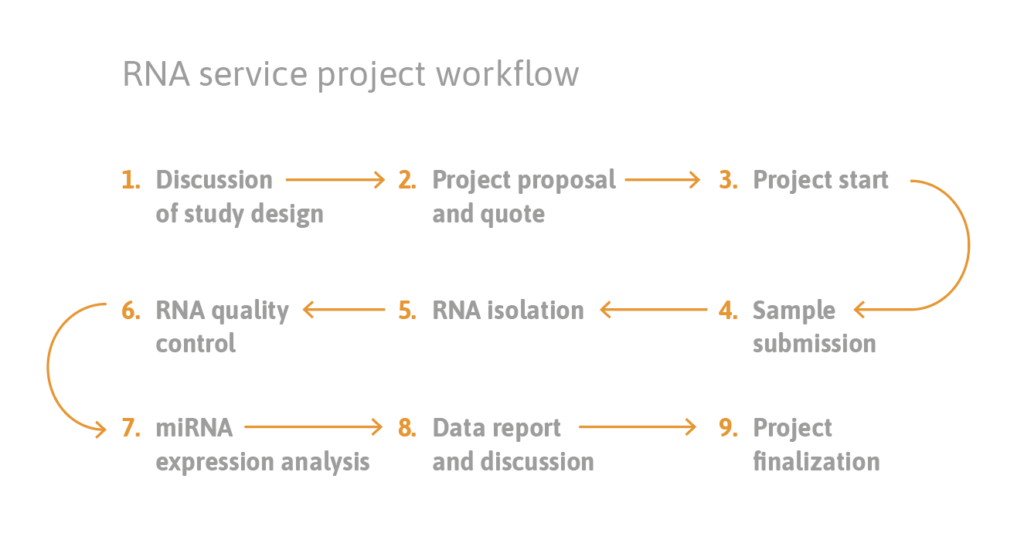
reference projects
Dexamethasone: a double-edged sword in the treatment of osteoarthritis
Tarasova K, Arteaga MB, Kidtiwong A et al.Sci Rep. 2025 Apr 7;15(1):11832. doi: 10.1038/s41598-025-96050-2
Sepsis induces heterogeneous transcription of coagulation- and inflammation-associated genes in renal microvasculature
Luxen M, Zwiers PJ, Jongmann RM et al. Thrombosis Research. March 18, 2024DOI:https://doi.org/10.1016/j.thromres.2024.03.014
Unique miRNome and transcriptome profiles underlie microvascular heterogeneity in mouse kidney
Luxen M, Zwiers PJ, Meester F et al. Am J Physiol Renal Physiol. 2023 Sep 1;325(3):F299-F316. doi: 10.1152/ajprenal.00005.2023. Epub 2023 Jul 6.
Comprehensive mRNA-sequencing-based characterization of three HEK-293 cell lines during an rAAV production process for Gene Therapy Applications
Pistek M., Kahlig CI., Hackl M. et al. Biotechnol J. 2023 May 16;e2200513. doi: 10.1002/biot.202200513
Transcriptomic landscapes of effective and failed liver regeneration in humans
Starlinger P., Brunnthaler L., McCabe C. et al. JHEP Reports, February 02, 2023 DOI:https://doi.org/10.1016/j.jhepr.2023.100683