One-stop-shop characterization of extracellular vesicles (EVs) and exosomes
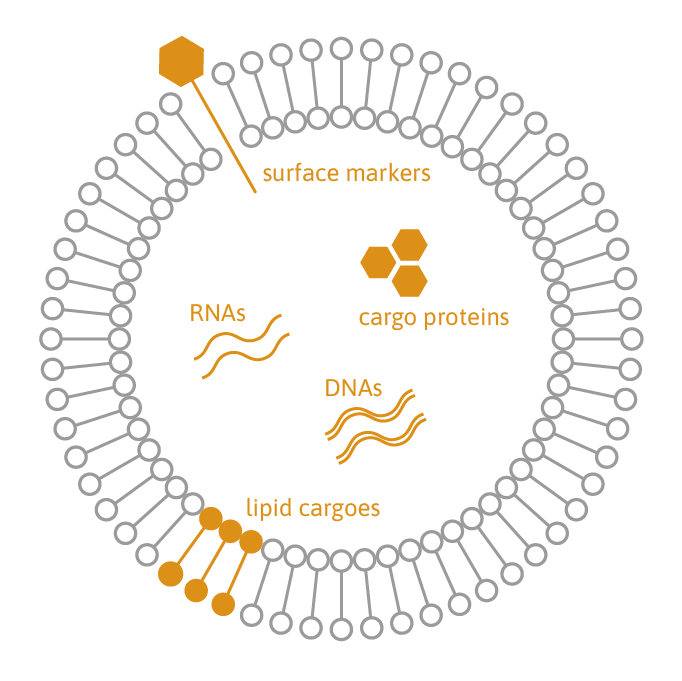
Extracellular vesicles (EVs) play a crucial role in cellular communication by transporting proteins, lipids, and RNA/DNA. They are emerging as valuable candidates for biomarkers and therapeutics in regenerative medicine, transplantation, cancer therapy, and immunotherapy.
TAmiRNA offers comprehensive EV characterization services, covering purification, quantification, and multi-omic analysis. We assess EV concentration, size distribution, and cargo composition, including nucleic acids (RNA/DNA), proteins, and lipids. Our proprietary small RNA sequencing protocol ensures absolute quantification of RNA molecules inside EVs using miND spike-in controls. Additionally, our optimized whole-transcriptome sequencing assay enables effective quantification of mRNA, long non-coding RNA (lncRNA), and circular RNA (circRNA) cargo in EVs.
All our services follow the best practices established by the International Society for Extracellular Vesicles (ISEV), ensuring high-quality and reproducible results.
Our EV services:
- EV/exosome purification from plasma, serum, urine, other biological matrices (e.g. stool), or conditioned cell culture media.
- Immunocapturing of EV populations based on surface markers.
- EV characterization (NTA, flow cytometry, and electron microscopy).
- RNA-sequencing analysis of microRNAs, mRNAs, lncRNAs and circRNAs.
- Mass-spec based proteomics for protein and lipid profiling.
benefits
EV purification: enrichment of EVs/exosomes can be performed using size exclusion chromatography (SEC), ultracentrifugation, and precipitation. SEC-based purification can deliver both EVs and protein complexes to study presence of RNA in both compartments in parallel.
EV characterization: we use Nanoparticle Tracking Analysis (NTA), cryo-electron microscopy, as well as multiplex flow cytometry to determine EV size, concentration, nucleic acid content (SYBR Gold), and surface epitopes (CD81, CD63, CD9). Upon request, we can develop assays for analyzing other surface epitopes.
Non-specific characterization of EV cargo: we apply highly sensitive methods for protein, dsDNA, and ssRNA quantification in EV preparations to characterize their protein and nucleic acid content. RNase/DNase/Protease treatments are used to reduce noise from surface decorations with RNA/DNA.
exRNA analysis in EVs: we have developed robust protocols for small RNAseq, RNAseq, and RT-qPCR analysis of EVs (and protein complexes) to perform highly quantitative analysis of microRNA, mRNA, and other non-coding RNAs.
Our experience: we have completed more RNA sequencing analyses of > 1000 EV samples obtained from liquid biopsies and conditioned media. We have developed protocols to purify EVs from stool samples and developed surface epitope staining protocols as well as protocols for staining intracellular RNA/DNA content using NTA analysis as a means of EV manufacturing quality control.
service requirements
We can purify EVs (and protein complexes) from
- biofluids such as (platelet-poor) plasma and urine,
- other biological matrices such as stool, synovial fluid, or milk,
- cell culture supernatant.
Sample volume is dependent on the type of biofluid, the required downstream analyses, and the concentration of EVs. Typically input volumes can be as low as 100 µl for plasma samples. Alternatively, we can process EV samples that have been generated by our customers as long as certain quality parameters can be assured. Get in touch with us to discuss your specific project details.
Comprehensive EV/Exosome characterization
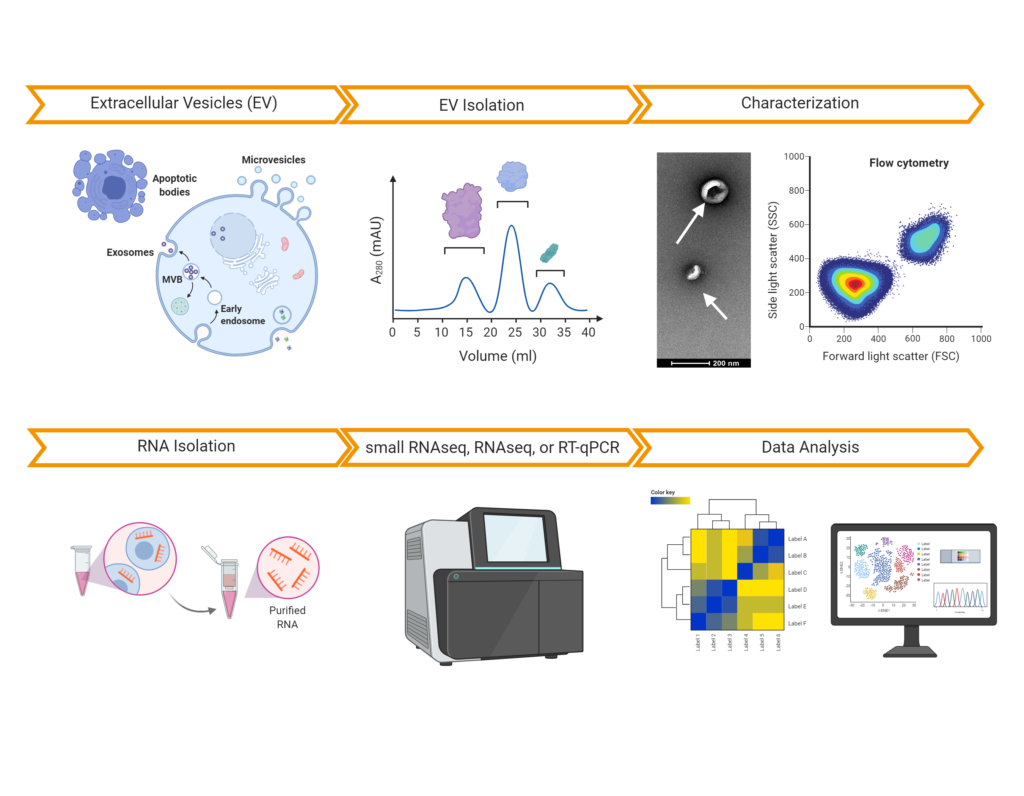
We have characterized >1000 EV / Exosome preparations from various sample types and species using NTA, RNA-sequencing, or proteomic analysis, enabling our customers and partners to increase the impact of their EV research projects. Here is a list of publications that have relied on our services:
reference projects
Extracellular Vesicle-Derived microRNA Crosstalk Between Equine Chondrocytes and Synoviocytes—An In Vitro Approach.
Castanheira C.I.G.D, Anderson R.J, Clarke E.J et al. Int. J. Mol. Sci. 2025, 26(7), 3353; https://doi.org/10.3390/ijms26073353
Snorkel-tag based affinity chromatography for recombinant extracellular vesicle purification. Bobbili MR, Görgens A, Yan Y et al. J Extracell Vesicles. 2024 Oct;13(10):e12523. doi: 10.1002/jev2.12523.
Neuron-Derived Extracellular Vesicles miRNA Profiles Identify Children Who Experience Adverse Events after Ketamine Administration for Procedural Sedation. Lucaf M, Bidoli C, Franzin M et al. Clin Pharmacol Ther. 2024 Aug 20. doi: 10.1002/cpt.3420
Identification of miRNAs Present in Cell- and Plasma-Derived Extracellular Vesicles—Possible Biomarkers of Colorectal Cancer. Lenart M, Sieminska I, Szatanek R et al. Cancers (Basel). 2024 Jul 5;16(13):2464. doi: 10.3390/cancers16132464.
Analysis of extracellular vesicle microRNA profiles reveals distinct blood and lymphatic endothelial cell origins. Pultar M, Oesterreicher J, Hartmann J et al. J of Extracellular Bio.2024;3:e134; https://doi.org/10.1002/jex2.134
Independent human mesenchymal stromal cell-derived extracellular vesicle preparations differentially attenuate symptoms in an advanced murine graft-versus-host disease model. Madel RJ, Börger V, Dittrich R et al. Cytotherapy. 2023 Aug;25(8):821-836. doi: 10.1016/j.jcyt.2023.03.008. Epub 2023 Apr 12.
Circulating endothelial extracellular vesicle signatures correspond with ICU requirement: an exploratory study in COVID-19 patients. Zipperle J, Oesterreicher J, Hackl M et al. Intensive Care Med Exp. 2023 Nov 30;11(1):85. doi: 10.1186/s40635-023-00567-7
Exosomal miRNAs from Prostate Cancer Impair Osteoblast Function in Mice. Furesi G, de Jesus Domingues AM, Alexopoulou D et al. Int J Mol Sci. 2022 Jan 24;23(3):1285. doi: 10.3390/ijms23031285.
Small non-coding RNA landscape of extracellular vesicles from a post-traumatic model of equine osteoarthritis Anderson JR., Jacobsen S., Walters M. et al. Front. Vet. Sci., 08 August 2022, Sec. Veterinary Regenerative Medicine, https://doi.org/10.3389/fvets.2022.901269
Functional repertoire of EV-associated miRNA profiles after lipoprotein depletion via ultracentrifugation and size exclusion chromatography from autologous blood products Otahal A., Kuten‑Pella O., Kramer K et al. Scientific Reports Volume 11, Article number: 5823 (2021).
SVF-derived extracellular vesicles carry characteristic miRNAs in lipedema. Priglinger E., Strohmeier K., Weigl M et al. Sci Rep. 2020 Apr 29;10(1):7211.doi: 10.1038/s41598-020-64215-w.