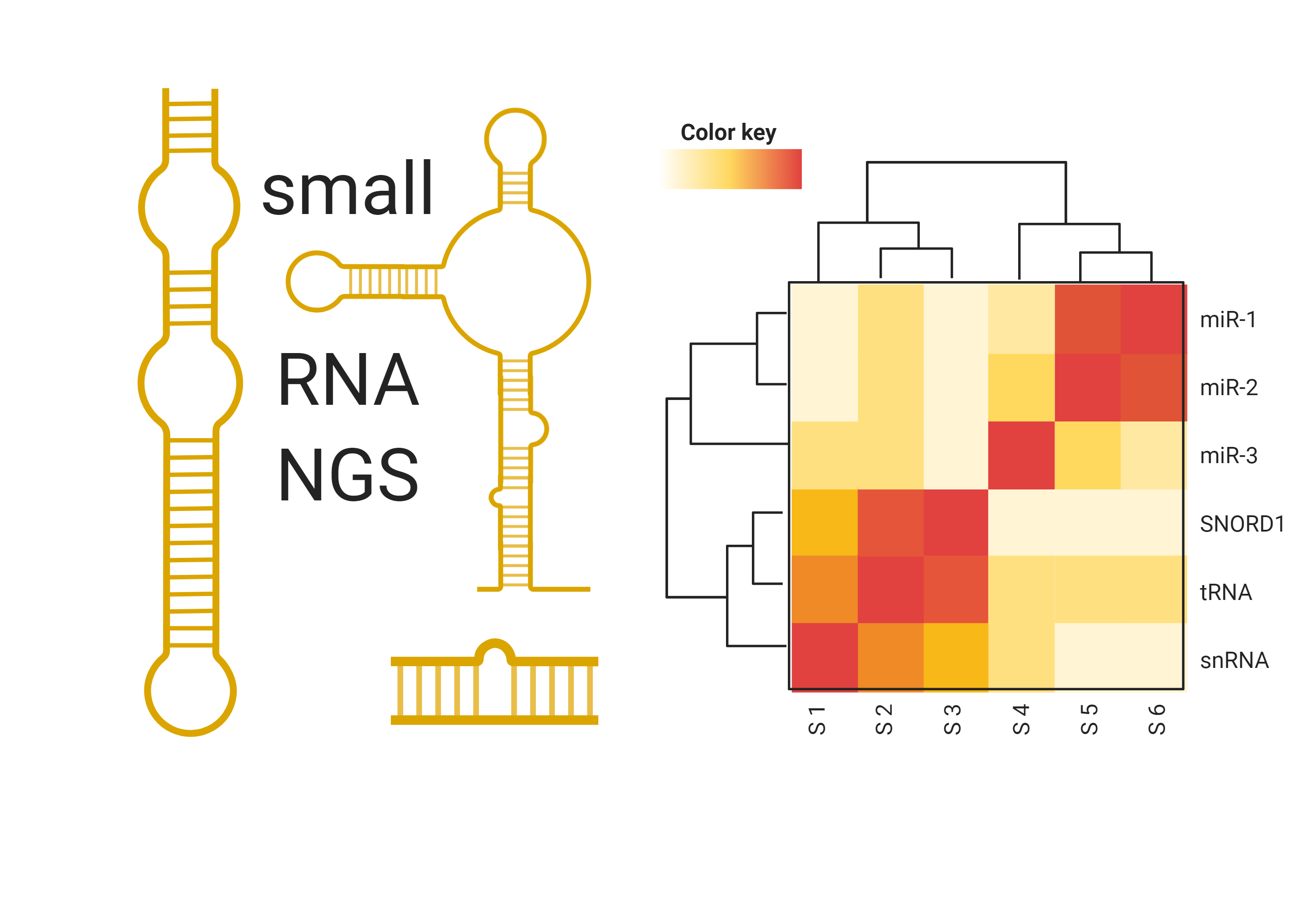
small RNA sequencing services
For untargeted genome-wide analysis of microRNAs and other small RNAs, next-generation sequencing (NGS) is the platform of choice.
At TAmiRNA we are fully dedicated to microRNA and small RNA NGS analysis. As we mainly work with extracellular RNA in biomarker research, we have long-lasting experience with very low-input RNA samples such as:
- biofluids (serum, plasma, platelet-poor plasma, urine, CSF, synovial fluid, milk, saliva, gut lumen, feces)
- microdissected tissue and cells,
- extracellular vesicles (EVs, exosomes) and protein complexes.
Our in-house developed miND® spike-ins enable absolute quantification and ensure the highest data quality, while RNA input can be reduced down to 1 ng total RNA or less.
Our NGS services go hand-in-hand with our
- RT-qPCR services to further verify and validate NGS findings,
- RNA-seq services to profile messenger RNA expression,
- Extracellular vesicle (EV) isolation and characterization services.
benefits
What we offer: our service includes total RNA extraction, small RNA sequencing library preparation, next-generation sequencing, data analysis, and reporting. Depending on customer experience we can focus on individual tasks or provide a full service.
Our promise: our small RNA sequencing workflow has undergone analytical validation. With best-in-class precision and accuracy, we can ensure the highest data quality. By using TAmiRNAs miND spike-ins we can transform relative counts into absolute concentrations (molecules/µL of input RNA).
Our experience: we have conducted small RNA sequencing analysis with samples from the following organisms: human, non-human primates, mouse, rat, hamster (CHO), pig, cow, horse, chicken, dogs, as well as C. elegans and drosophila. For other organisms data analysis can be implemented upon request. Browse our published reference projects (below) for more information on previous projects.
Your data: we provide interactive reports (see video above) that enable you to access and download your data so that it can be directly integrated into reports, publications, and presentations.
Your flexibility: we offer MiSeq, iSeq, NextSeq550, NextSeq1000/2000 and the NovaSeq. Thus, we manage to find a cost-effective solution for small and large projects.
sample requirements
Cells: we can go as low as 10-50 cells
Tissues: we can process everything from laser microdissected tissue compartments of just 10,000 µm2 up to frozen or fixated tissue blocks. Learn more about our workflow for space-resolved RNA profiling.
Biofluids: serum, plasma, urine, saliva, CSF, milk, synovial fluid. The standard input is 100-200 µl but can be optimized to meet your restrictions.
Extracellular vesicles: characterization of EV RNA cargo can be performed by NGS or RT-qPCR. Due to the heterogeneity of methods that are used for EV purification, we prefer to discuss your project individually.
Testimonials
“Since our first contact in July 2016, many projects have been successfully conducted with TAmiRNA, leading to peer-reviewed articles and novel findings on biomarkers. This achievement has been possible because of the expertise and support of TAmiRNA together with the speed of the service.” .
“”I worked with Matthias Hackl and his TAmiRNA team for two projects. I was very happy with my results and found Matthias and his team very helpful and accommodating of our timeline for assay completion. Matthias provided invaluable assistance in the interpretation of the microRNA results. I highly recommend using TAmiRNA services for microRNA analysis.”.
“They have helped us getting the best from out data analysis and interpretation. Their flexibility and experience makes them an excellent partner of choice”.
service description
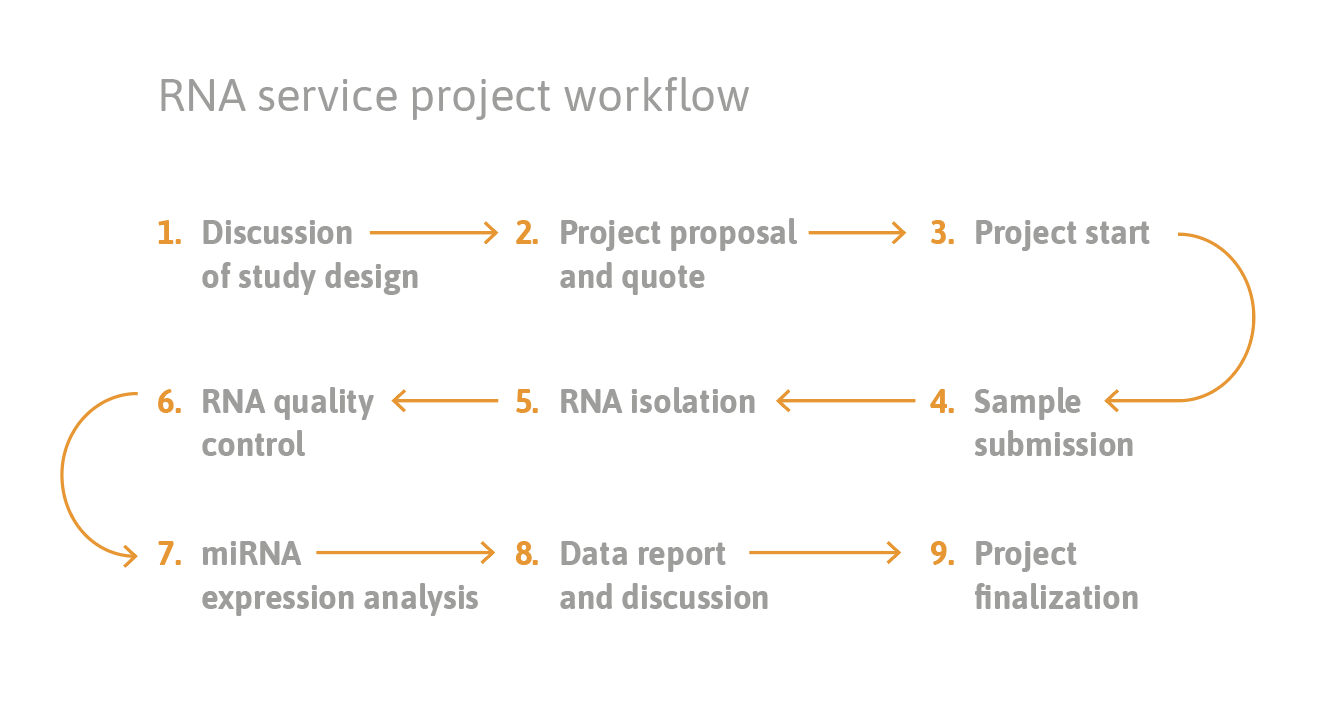
Small RNA sequencing includes several steps, which can all be performed at TAmiRNA:
- Total RNA extraction: we have protocols for cells, tissues, biofluids and EVs. Leftover total RNA can be returned to customers for follow-up experiments.
- Total RNA quality control: concentration and integrity using highly sensitive and specific fluorescence based methods
- small RNA library preparation: we prepare high quality libraries using a circularization protocol. This reduce adapter ligation bias compared to other methods. In addition, we have developed a proprietary spike-in calibrator that is added to the library preparation and enables absolute quantification.
- NGS analysis: using a wide-range of Illumina instruments (HiSeq, NextSeq, NovaSeq) we can pick the most efficient solution for each project.
- Data analysis: we provide everything from raw data to fully analyzed data reports depending on customer needs.
reference projects
Aged-vascular niche hinders osteogenesis of mesenchymal stem cells through paracrine repression of Wnt-axis
Fleischhacker V, Milosic F, Bricelj M et al. Aging Cell 2024, https://doi.org/10.1111/acel.14139
MicroRNA-30d-5p—A Potential New Therapeutic Target for Prevention of Ischemic Cardiomyopathy after Myocardial Infarction
Boxhammer E, Paar V, Wernly B et al. Cells 2023, 12(19), 2369; https://doi.org/10.3390/cells12192369
Unique miRNome and transcriptome profiles underlie microvascular heterogeneity in mouse kidney
Luxen M, Zwiers PJ, Meester F et al. Am J Physiol Renal Physiol. 2023 Sep 1;325(3):F299-F316. doi: 10.1152/ajprenal.00005.2023. Epub 2023 Jul 6.
Investigation of MicroRNA Biomarkers in Equine Distal Interphalangeal Joint Osteoarthritis
Baker ME., Lee S., Clinton M. et al. Int J Mol Sci 2022 Dec 8;23(24):15526. doi: 10.3390/ijms232415526.
Modulation of Host-Parasite Interactions with Small Molecules Targeting Schistosoma mansoni microRNAs
Hamway Y., Zimmermann K., Blommers MJJ. et al. ACS Infect Dis. 2022 Oct 14;8(10):2028-2034. doi: 10.1021/acsinfecdis.2c00360. Epub 2022 Sep 13.
Comprehensive Characterization of Platelet-Enriched MicroRNAs as Biomarkers of Platelet Activation
Krammer TL., Zeibig S., Schrottmaier WC. et al. Cells 2022 Apr 7;11(8):1254. doi: 10.3390/cells11081254.
The role of extracellular vesicle miRNAs and tRNAs in synovial fibroblast senescence
Wijesinghe SN., Anderson J., Brown TJ. et al. Front Mol Biosci. 2022 Sep 23;9:971621. doi: 10.3389/fmolb.2022.971621. eCollection 2022.
Small non-coding RNA landscape of extracellular vesicles from a post-traumatic model of equine osteoarthritis
Anderson JR., Jacobsen S., Walters M. et al. Front Vet Sci. 2022 Aug 8;9:901269. doi: 10.3389/fvets.2022.901269. eCollection 2022.
MicroRNA Expression Profiling in Porcine Liver, Jejunum and Serum upon Dietary DON Exposure Reveals Candidate Toxicity Biomarkers
Segura-Wang M. Grenier B., Ilic S. et al. Int J Mol Sci. 2021 Nov 7;22(21):12043. doi: 10.3390/ijms222112043./em>
Next-generation sequencing analysis of circulating micro-RNA expression in response to parabolic flight as a spaceflight analogue
Jirak P., Wernly B., Lichtenauer M. et al. npj Microgravity volume 6, Article number: 31 (2020)
MicroRNA Expression Profile Changes after Cardiopulmonary Bypass and Ischemia/Reperfusion-Injury in a Porcine Model of Cardioplegic Arrest.
Kiss A., Heber S., Kramer AM. et al. Diagnostics (Basel). 2020 Apr 21;10(4):240. doi: 10.3390/diagnostics10040240.
Anti-CD3 Antibody Treatment Reduces Scar Formation in a Rat Model of Myocardial Infarction.
Wernly B., Paar V., Aigner A. et al Cells. 2020 Jan 25;9(2):295. doi: 10.3390/cells9020295.
Combined proteomics/miRNomics of dendritic cell immunotherapy-treated glioblastoma patients as a screening for survival-associated factors.
Erhart F., Hackl M., Hahne H. et al. NPJ Vaccines. 2020 Jan 16;5:5. doi: 10.1038/s41541-019-0149-x. eCollection 2020.
MicroRNA levels in bone and blood change during bisphosphonate and teriparatide therapy in an animal model of postmenopausal osteoporosis.
Kocijan R., Weigl M., Skalicky S. et al. Bone. 2020 Feb;131:115104. doi: 10.1016/j.bone.2019.115104. Epub 2019 Nov 1.
MicroRNAs in porcine uterus and serum are affected by zearalenone and represent a new target for mycotoxin biomarker discovery.
Grenier B., Hackl M., Skalicky S. et al. Sci Rep. 2019 Jun 28;9(1):9408. doi: 10.1038/s41598-019-45784-x.
Biotransformation of the Mycotoxin Zearalenone to its Metabolites Hydrolyzed Zearalenone (HZEN) and Decarboxylated Hydrolyzed Zearalenone (DHZEN) Diminishes its Estrogenicity In Vitro and In Vivo.
Fruhauf S, Novak B, Nagl V et al. Toxins (Basel). 2019 Aug 20;11(8). pii: E481. doi: 10.3390/toxins11080481
Predicting Postoperative Liver Dysfunction Based on Blood-Derived MicroRNA Signatures.
Starlinger P., Hackl H., Pereyra D. et al. Hepatology. 2019 Jun;69(6):2636-2651. doi: 10.1002/hep.30572. Epub 2019 Apr 10.
Small extracellular vesicles and their miRNA cargo are anti-apoptotic members of the senescence-associated secretory phenotype.
Terlecki-Zaniewicz L., Lämmermann I., Latreille J. et al. Aging (Albany NY). 2018 May 19;10(5):1103-1132. doi: 10.18632/aging.101452.
A signature of 12 microRNAs is robustly associated with growth rate in a variety of CHO cell lines.
Klanert G., Jadhav V., Shanmukam V. et al. J Biotechnol. 2016 Oct 10;235:150-61. doi: 10.1016/j.jbiotec.2016.03.022. Epub 2016 Mar 16.