miND® Spike-In fulfills two primary purposes:
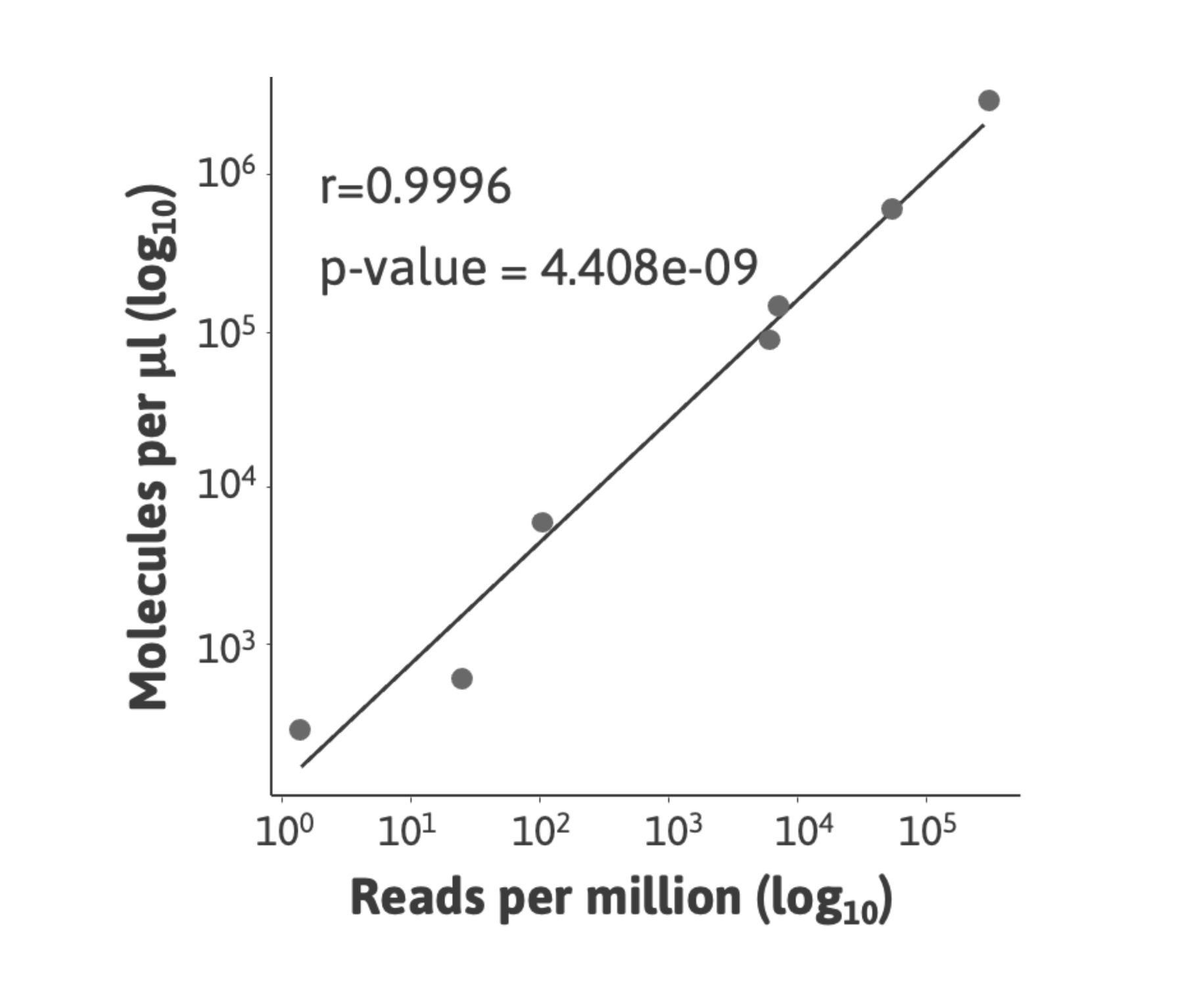
1. Quality control: confirm the dynamic range and quantitativeness of your small RNA sequencing experiments.
2. Absolute normalization: convert read counts to copies/µl total RNA and reduce bias originating from variation in RNA composition between samples.
miND® Spike-In design and usability:
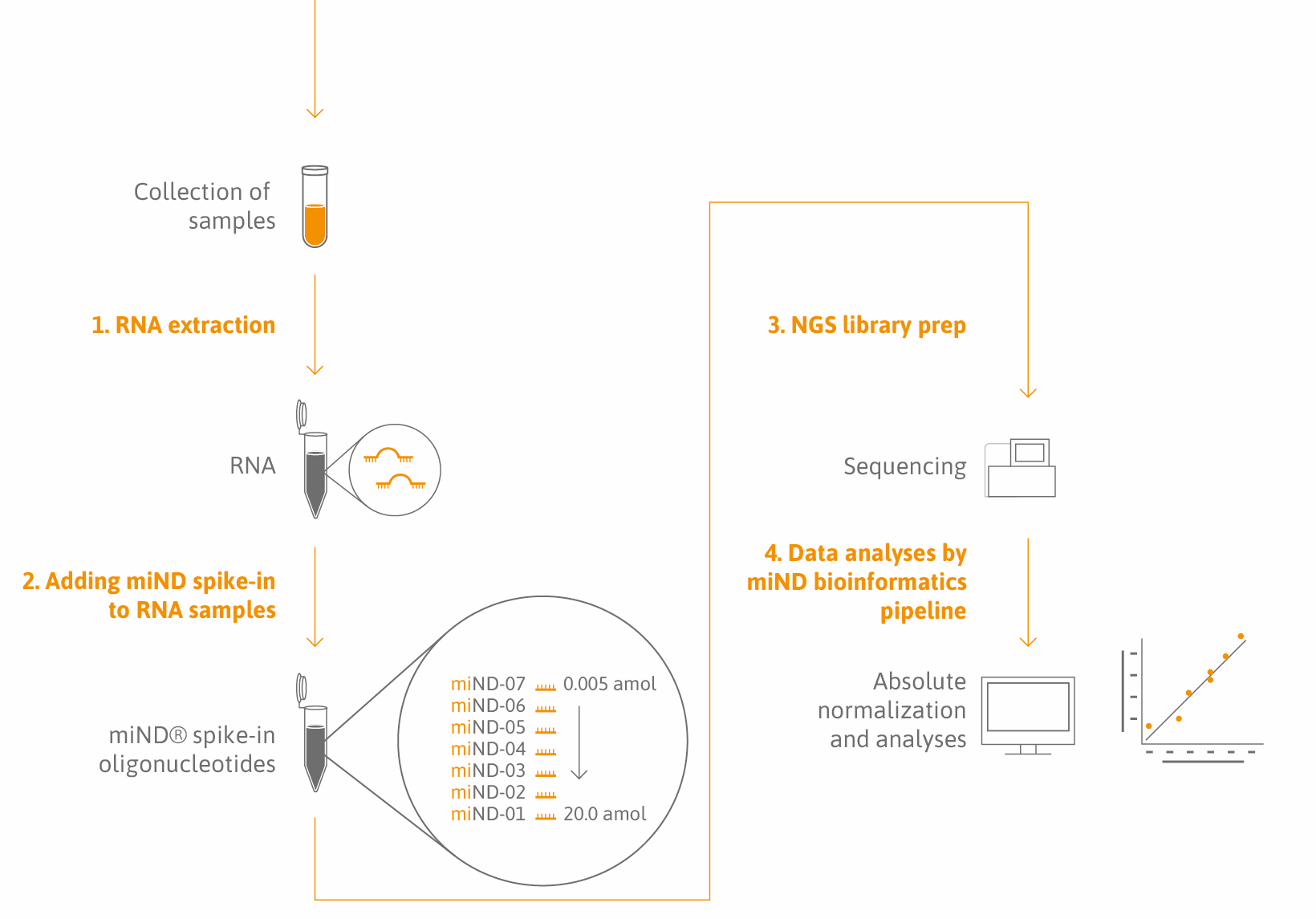
miND® Spike-Ins are a proprietary oligonucleotide mix with unique design features (Lutzmayer et al.). They are added to your total RNA samples prior to microRNA or small RNA sequencing analysis. The spike-in sequence composition and concentration range have been optimized to be compatible with almost any species as well as a broad range of sample types including biofluids (serum, plasma, urine, CSF, synovial fluid), cells, tissues, extracellular vesicles, and non-vesicular fractions (Khamina et al.).
benefits
The challenge: small RNA-sequencing, especially for challenging input samples such as biofluids, exosomes, or low cell numbers, is prone to sequencing bias. In addition, the comparison of microRNAs and other small RNAs between sample types with differing RNA compositions is skewed based on the assumption of a constant amount of small RNAs per sample (the underlying hypothesis for reads per million (RPM) normalization).
Our solution: miND® Spike-Ins are a novel quality control parameter and normalizer that consists of seven oligonucleotides, each characterized by a unique core sequence flanked by 4 randomized nucleotides. miND® Spike-Ins are provided in a specific ratio to cover the broad concentration range of endogenous small RNAs.
Simplicity: miND® Spike-Ins come as ready-to-use reagents. No dilution, mixing, or change in your workflow is required – miND® spike-ins are directly added to your RNA sample and used for NGS library preparation.
Sample-to-insight: our public data analysis pipeline was specifically developed to convert small RNA-sequencing raw data into a simple but comprehensive report providing full access to your data in the conventional way (RPM, read count) as well as in absolute concentrations. On top of that, we have added several unsupervised and supervised analyses and QC parameters to the report.
Not familiar with small RNA-sequencing?
No problem – the entire miND small RNA-seq workflow is provided as a service by TAmiRNA – request a quotation.
Curious to learn more about miND® Spike-In?
Sign up for our newsletter and get the exemplary miND® NGS report
price list
| product number | product | size/formulation | shipment conditions | price * | product information | order |
|---|---|---|---|---|---|---|
| KT-041-MIND-250 | miND spike in | 250 reactions – lyophilized | room temperature | € 750 | product sheet / manual | request quote |
| KT-041-MIND-48 | miND spike in | 48 reactions – resuspended | dry ice | € 250 | product sheet / manual | request quote |
* excluding shipment
product description
- 7 unique core sequences mixed in a specific proportions to cover a wide range of endogenous microRNAs
- Each unique core sequence consists of 13 nt that doesn’t match the genome of interest
- The core sequence is flanked by a set of 4 randomized nucleotides on both 5’ and 3’ ends
- Each of these 13 nt unique core sequences can be represented by up to 65,536 possible 21 nt sequences
- The miND spike-in contains up to 458,752 unique oligonucleotides
key publications and reference projects
Identification of miRNAs Present in Cell- and Plasma-Derived Extracellular Vesicles—Possible Biomarkers of Colorectal Cancer. Lenart M, Sieminska I, Szatanek R et al. Cancers (Basel). 2024 Jul 5;16(13):2464. doi: 10.3390/cancers16132464.
Analysis of extracellular vesicle microRNA profiles reveals distinct blood and lymphatic endothelial cell origins. Pultar M, Oesterreicher J, Hartmann J et al. J of Extracellular Bio.2024;3:e134; https://doi.org/10.1002/jex2.134
A MicroRNA Next-Generation-Sequencing Discovery Assay (miND) for Genome-Scale Analysis and Absolute Quantitation of Circulating MicroRNA Biomarkers Khamina K, Diendorfer A, Skalicky S, et al. Int. J. Mol. Sci. 2022, 23(3), 1226; https://doi.org/10.3390/ijms23031226
miND (miRNA NGS Discovery pipeline): a small RNA-seq analysis pipeline and report generator for microRNA biomarker discovery studies Diendorfer A, Khamina K, Pultar M and Hackl M. F1000Research 2022, 11:233, 1226; https://doi.org/10.12688/f1000research.94159.1
MicroRNA-30d-5p—A Potential New Therapeutic Target for Prevention of Ischemic Cardiomyopathy after Myocardial Infarction
Boxhammer E, Paar V, Wernly B et al. Cells 2023, 12(19), 2369; https://doi.org/10.3390/cells12192369
Unique miRNome and transcriptome profiles underlie microvascular heterogeneity in mouse kidney
Luxen M, Zwiers PJ, Meester F et al. Am J Physiol Renal Physiol. 2023 Sep 1;325(3):F299-F316. doi: 10.1152/ajprenal.00005.2023. Epub 2023 Jul 6.
Investigation of MicroRNA Biomarkers in Equine Distal Interphalangeal Joint Osteoarthritis
Baker ME., Lee S., Clinton M. et al. Int J Mol Sci 2022 Dec 8;23(24):15526. doi: 10.3390/ijms232415526.
Modulation of Host-Parasite Interactions with Small Molecules Targeting Schistosoma mansoni microRNAs
Hamway Y., Zimmermann K., Blommers MJJ. et al. ACS Infect Dis. 2022 Oct 14;8(10):2028-2034. doi: 10.1021/acsinfecdis.2c00360. Epub 2022 Sep 13.
Comprehensive Characterization of Platelet-Enriched MicroRNAs as Biomarkers of Platelet Activation
Krammer TL., Zeibig S., Schrottmaier WC. et al. Cells 2022 Apr 7;11(8):1254. doi: 10.3390/cells11081254.
The role of extracellular vesicle miRNAs and tRNAs in synovial fibroblast senescence
Wijesinghe SN., Anderson J., Brown TJ. et al. Front Mol Biosci. 2022 Sep 23;9:971621. doi: 10.3389/fmolb.2022.971621. eCollection 2022.
Small non-coding RNA landscape of extracellular vesicles from a post-traumatic model of equine osteoarthritis
Anderson JR., Jacobsen S., Walters M. et al. Front Vet Sci. 2022 Aug 8;9:901269. doi: 10.3389/fvets.2022.901269. eCollection 2022.
MicroRNA Expression Profiling in Porcine Liver, Jejunum and Serum upon Dietary DON Exposure Reveals Candidate Toxicity Biomarkers
Segura-Wang M. Grenier B., Ilic S. et al. Int J Mol Sci. 2021 Nov 7;22(21):12043. doi: 10.3390/ijms222112043./em>
Novel small RNA spike-in oligonucleotides enable absolute normalization of small RNA-Seq data Lutzmayer S, Enugutti B and Nodine MD. Sci Rep. 2017 Jul 19;7(1):5913. doi: 10.1038/s41598-017-06174-3.
Small non-coding RNA landscape of extracellular vesicles from a post-traumatic model of equine osteoarthritis Anderson JR., Jacobsen S., Walters M. et al. Front. Vet. Sci., 08 August 2022, Sec. Veterinary Regenerative Medicine, https://doi.org/10.3389/fvets.2022.901269