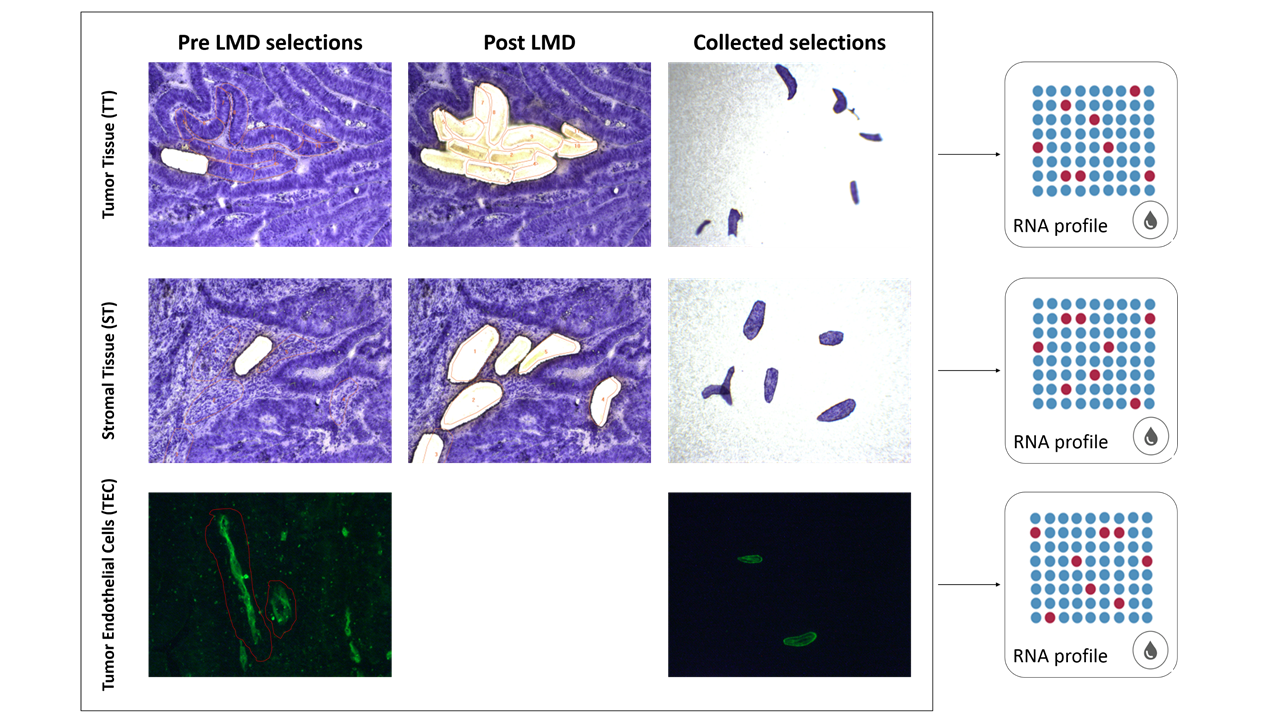
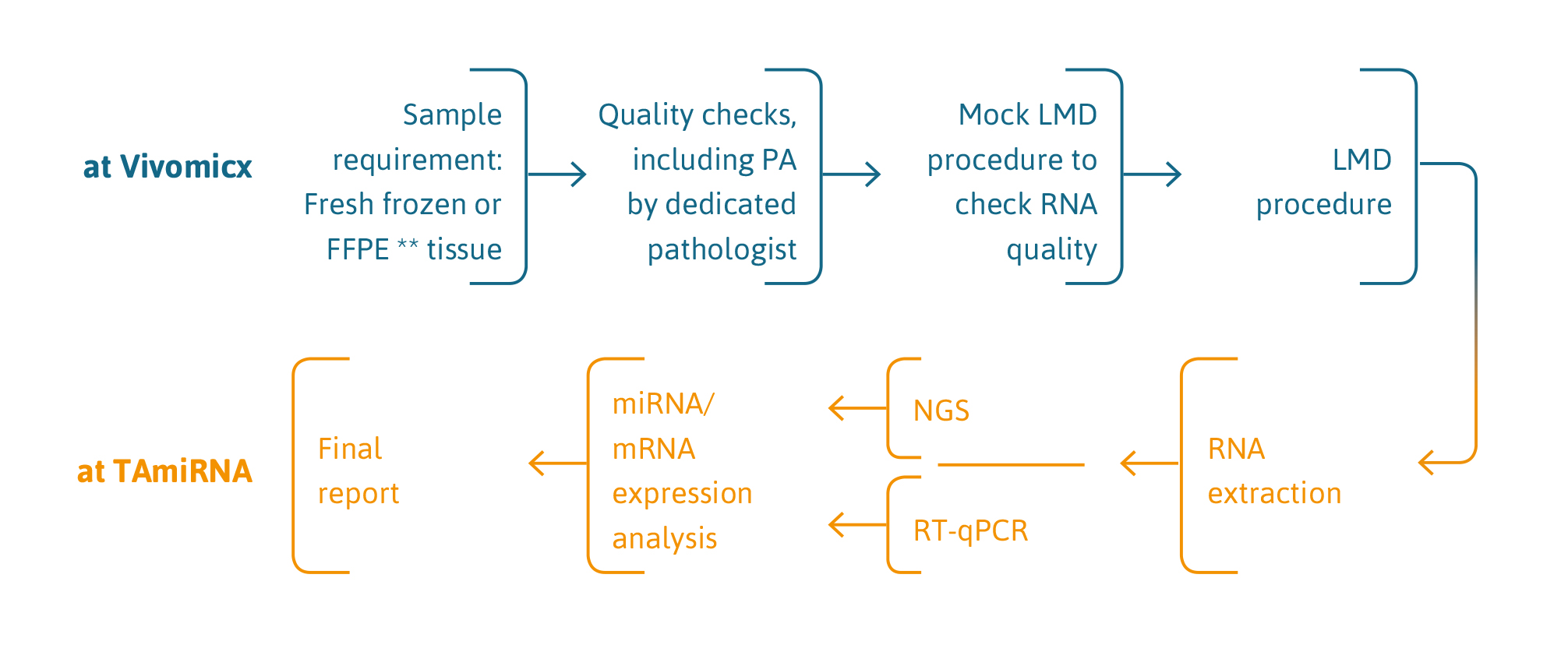
Cell-type specific and space-resolved RNA profiling in complex tissues
Healthy and malignant tissues are composed of multiple cell types: tumor and stromal cells, microvasculature and other specialized cell types (e.g., Tumor Infiltrating Lymphocytes). Therefore, the analysis of whole tumors results in a mixture of signals with unclear origin. Deconvolution of tissue heterogeneity and sensitive analytical methods are essential to obtain interpretable results from tissue analysis.
TAmiRNA and Dutch company Vivomicx have combined their respective technologies of high sensitivity gene expression profiling (RT-qPCR and smallRNAseq/RNAseq) and laser microdissection to enable cell-type specific and space-resolved RNA expression analysis in complex tissues.
This technology provides more accurate information on cellular states during onset and progression of diseases to accelerate drug and biomarker discovery.
benefits
Space-resolved and cell-type-specific gene expression analysis provide deep insights into disease mechanisms.
The high signal-to-noise ratio of space-resolved gene expression analysis enables the identification of novel drug and biomarker candidates.
The effect of drug-target engagement can be monitored in situ at the level of individual cell types, providing accurate insights into drug mode-of-action.
The combination of laser microdissection (LMD) and high sensitivity gene expression profiling can be applied to almost any biological tissue sample.
Every project is different and has specific requirements. Experts at Vivomicx and TAmiRNA have a combined experience of >15 years and are committed to finding customer-tailored solutions.
sample requirements
Laser microdissection requires preserved tissues such as fresh frozen or fixated tissue.
We acknowledge that every LMD project is different. Our technical experts will discuss sample requirements at the beginning of each project and pilot experiments can be performed to optimize performance.
service description
Using this protocol, TAmiRNA & Vivomicx have been able to:
- Demonstrate significant differences in RNA expression between whole tumor and subsets of cells within the tumor (Skalicky et al. Biotechniques, 2019).
- Unravel small RNA regulation in the tumor, stroma, and tumor endothelial cells from colorectal carcinoma samples.
- Catalog small RNA expression patterns in kidney compartments: glomeruli, tubules, and peritubular capillaries.
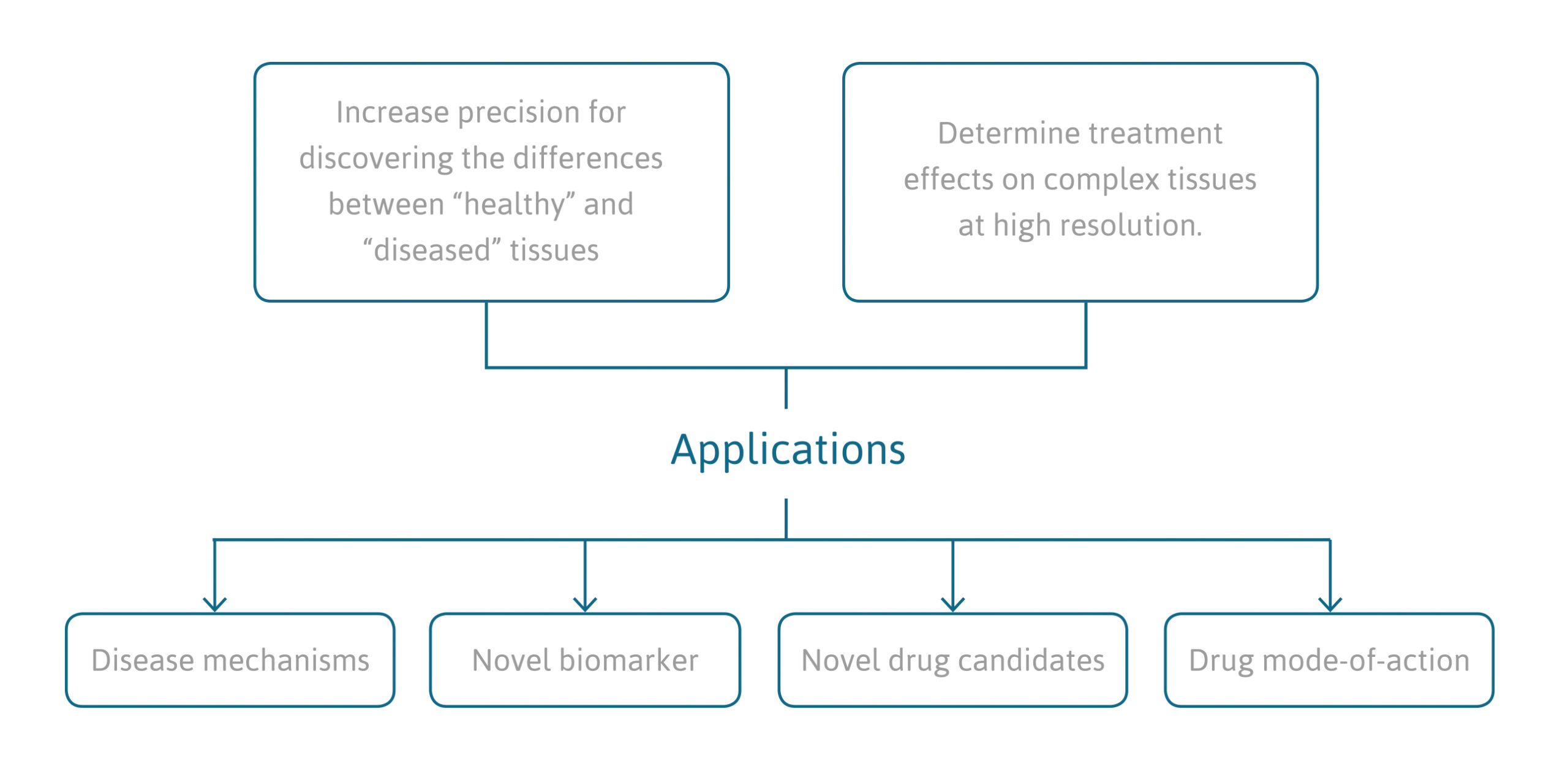
application of this technology
- Discovery of novel drug and biomarker candidates through space-resolved analysis of diseased versus healthy tissue samples.
- Identification of drug mode of actions by analyzing the effects of therapeutic interventions on gene expression in specific cell-types within the complexity of a tissue.
- Quantify drug-target engagement in specific cell types in complex tissue through the screening of downstream effects on gene expression level.
- Explore disease mechanisms at highest resolution and improved signal-to-noise ratio.
reference projects
Sepsis induces heterogeneous transcription of coagulation- and inflammation-associated genes in renal microvasculature
Luxen M, Zwiers PJ, Jongmann RM et al. Thrombosis Research. March 18, 2024DOI:https://doi.org/10.1016/j.thromres.2024.03.014
Unique miRNome and transcriptome profiles underlie microvascular heterogeneity in mouse kidney
Luxen M, Zwiers PJ, Meester F et al. Am J Physiol Renal Physiol. 2023 Sep 1;325(3):F299-F316. doi: 10.1152/ajprenal.00005.2023. Epub 2023 Jul 6.
Combining laser microdissection and microRNA expression profiling to unmask microRNA signatures in complex tissues
Skalicky S, Zwiers PJ, Kuiper T et al. Biotechniques. 2019 Oct 17. doi: 10.2144/btn-2019-0032. Epub 2019 Oct 17.
MicroRNA-126 contributes to renal microvascular heterogeneity of VCAM-1 protein expression in acute inflammation
2012 Jun 15;302(12):F1630-9.